Sleep Health and Lifestyle#
Author: Xidan Liao
Course Project, UC Irvine, Math 10, F23
Introduction#
Project#
This project attempts to analyze and predict people’s sleep status through their daily habits and lifestyles. The variables include “Quality of Sleep”, “Physical Activity Level”, “Stress Level” and so on. Our main topics include the length of sleep and the presence of sleep disorders.
Dataset Columns#
Person ID: An identifier for each individual.Gender:The gender of the person (Male/Female).Age: The age of the person in years.Occupation: The occupation or profession of the person.Sleep Duration (hours): The number of hours the person sleeps per day.Quality of Sleep (scale: 1-10): A subjective rating of the quality of sleep, ranging from 1 to 10.Physical Activity Level (minutes/day): The number of minutes the person engages in physical activity daily.Stress Level (scale: 1-10): A subjective rating of the stress level experienced by the person, ranging from 1 to 10.BMI Category: The BMI category of the person (e.g., Underweight, Normal, Overweight).Blood Pressure (systolic/diastolic): The blood pressure measurement of the person, indicated as systolic pressure over diastolic pressure.Heart Rate (bpm): The resting heart rate of the person in beats per minute.Daily Steps: The number of steps the person takes per day.Sleep Disorder: The presence or absence of a sleep disorder in the person (None, Insomnia, Sleep Apnea).
Details about Sleep Disorder Column:
None: The individual does not exhibit any specific sleep disorder.Insomnia: The individual experiences difficulty falling asleep or staying asleep, leading to inadequate or poor-quality sleep.Sleep Apnea: The individual suffers from pauses in breathing during sleep, resulting in disrupted sleep patterns and potential health risks.
Dataset preprocessing#
Importing Dataset#
import pandas as pd
import numpy as np
import seaborn as sns
import altair as alt
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import PolynomialFeatures
from sklearn.metrics import mean_squared_error
from sklearn.tree import DecisionTreeClassifier
from sklearn.tree import plot_tree
from sklearn.ensemble import RandomForestClassifier
df = pd.read_csv("Sleep_health_and_lifestyle_dataset.csv")
df.shape
(374, 13)
df
| Person ID | Gender | Age | Occupation | Sleep Duration | Quality of Sleep | Physical Activity Level | Stress Level | BMI Category | Blood Pressure | Heart Rate | Daily Steps | Sleep Disorder | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | Male | 27 | Software Engineer | 6.1 | 6 | 42 | 6 | Overweight | 126/83 | 77 | 4200 | None |
| 1 | 2 | Male | 28 | Doctor | 6.2 | 6 | 60 | 8 | Normal | 125/80 | 75 | 10000 | None |
| 2 | 3 | Male | 28 | Doctor | 6.2 | 6 | 60 | 8 | Normal | 125/80 | 75 | 10000 | None |
| 3 | 4 | Male | 28 | Sales Representative | 5.9 | 4 | 30 | 8 | Obese | 140/90 | 85 | 3000 | Sleep Apnea |
| 4 | 5 | Male | 28 | Sales Representative | 5.9 | 4 | 30 | 8 | Obese | 140/90 | 85 | 3000 | Sleep Apnea |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 369 | 370 | Female | 59 | Nurse | 8.1 | 9 | 75 | 3 | Overweight | 140/95 | 68 | 7000 | Sleep Apnea |
| 370 | 371 | Female | 59 | Nurse | 8.0 | 9 | 75 | 3 | Overweight | 140/95 | 68 | 7000 | Sleep Apnea |
| 371 | 372 | Female | 59 | Nurse | 8.1 | 9 | 75 | 3 | Overweight | 140/95 | 68 | 7000 | Sleep Apnea |
| 372 | 373 | Female | 59 | Nurse | 8.1 | 9 | 75 | 3 | Overweight | 140/95 | 68 | 7000 | Sleep Apnea |
| 373 | 374 | Female | 59 | Nurse | 8.1 | 9 | 75 | 3 | Overweight | 140/95 | 68 | 7000 | Sleep Apnea |
374 rows × 13 columns
Checking for missing values#
missing_values = df.isnull().sum()
missing_values
Person ID 0
Gender 0
Age 0
Occupation 0
Sleep Duration 0
Quality of Sleep 0
Physical Activity Level 0
Stress Level 0
BMI Category 0
Blood Pressure 0
Heart Rate 0
Daily Steps 0
Sleep Disorder 0
dtype: int64
There is no missing value.
Converting to category type#
df.dtypes
Person ID int64
Gender object
Age int64
Occupation object
Sleep Duration float64
Quality of Sleep int64
Physical Activity Level int64
Stress Level int64
BMI Category object
Blood Pressure object
Heart Rate int64
Daily Steps int64
Sleep Disorder object
dtype: object
df['Gender_cat'] = df['Gender'].astype('category').cat.codes
df['Occupation_cat'] = df['Occupation'].astype('category').cat.codes
df['BMI Category_cat'] = df['BMI Category'].astype('category').cat.codes
df['Sleep Disorder_cat'] = df['Sleep Disorder'].astype('category').cat.codes
df
| Person ID | Gender | Age | Occupation | Sleep Duration | Quality of Sleep | Physical Activity Level | Stress Level | BMI Category | Blood Pressure | Heart Rate | Daily Steps | Sleep Disorder | Gender_cat | Occupation_cat | BMI Category_cat | Sleep Disorder_cat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | Male | 27 | Software Engineer | 6.1 | 6 | 42 | 6 | Overweight | 126/83 | 77 | 4200 | None | 1 | 9 | 3 | 1 |
| 1 | 2 | Male | 28 | Doctor | 6.2 | 6 | 60 | 8 | Normal | 125/80 | 75 | 10000 | None | 1 | 1 | 0 | 1 |
| 2 | 3 | Male | 28 | Doctor | 6.2 | 6 | 60 | 8 | Normal | 125/80 | 75 | 10000 | None | 1 | 1 | 0 | 1 |
| 3 | 4 | Male | 28 | Sales Representative | 5.9 | 4 | 30 | 8 | Obese | 140/90 | 85 | 3000 | Sleep Apnea | 1 | 6 | 2 | 2 |
| 4 | 5 | Male | 28 | Sales Representative | 5.9 | 4 | 30 | 8 | Obese | 140/90 | 85 | 3000 | Sleep Apnea | 1 | 6 | 2 | 2 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 369 | 370 | Female | 59 | Nurse | 8.1 | 9 | 75 | 3 | Overweight | 140/95 | 68 | 7000 | Sleep Apnea | 0 | 5 | 3 | 2 |
| 370 | 371 | Female | 59 | Nurse | 8.0 | 9 | 75 | 3 | Overweight | 140/95 | 68 | 7000 | Sleep Apnea | 0 | 5 | 3 | 2 |
| 371 | 372 | Female | 59 | Nurse | 8.1 | 9 | 75 | 3 | Overweight | 140/95 | 68 | 7000 | Sleep Apnea | 0 | 5 | 3 | 2 |
| 372 | 373 | Female | 59 | Nurse | 8.1 | 9 | 75 | 3 | Overweight | 140/95 | 68 | 7000 | Sleep Apnea | 0 | 5 | 3 | 2 |
| 373 | 374 | Female | 59 | Nurse | 8.1 | 9 | 75 | 3 | Overweight | 140/95 | 68 | 7000 | Sleep Apnea | 0 | 5 | 3 | 2 |
374 rows × 17 columns
Q1: Prediction of Sleep Duration#
Visualization#
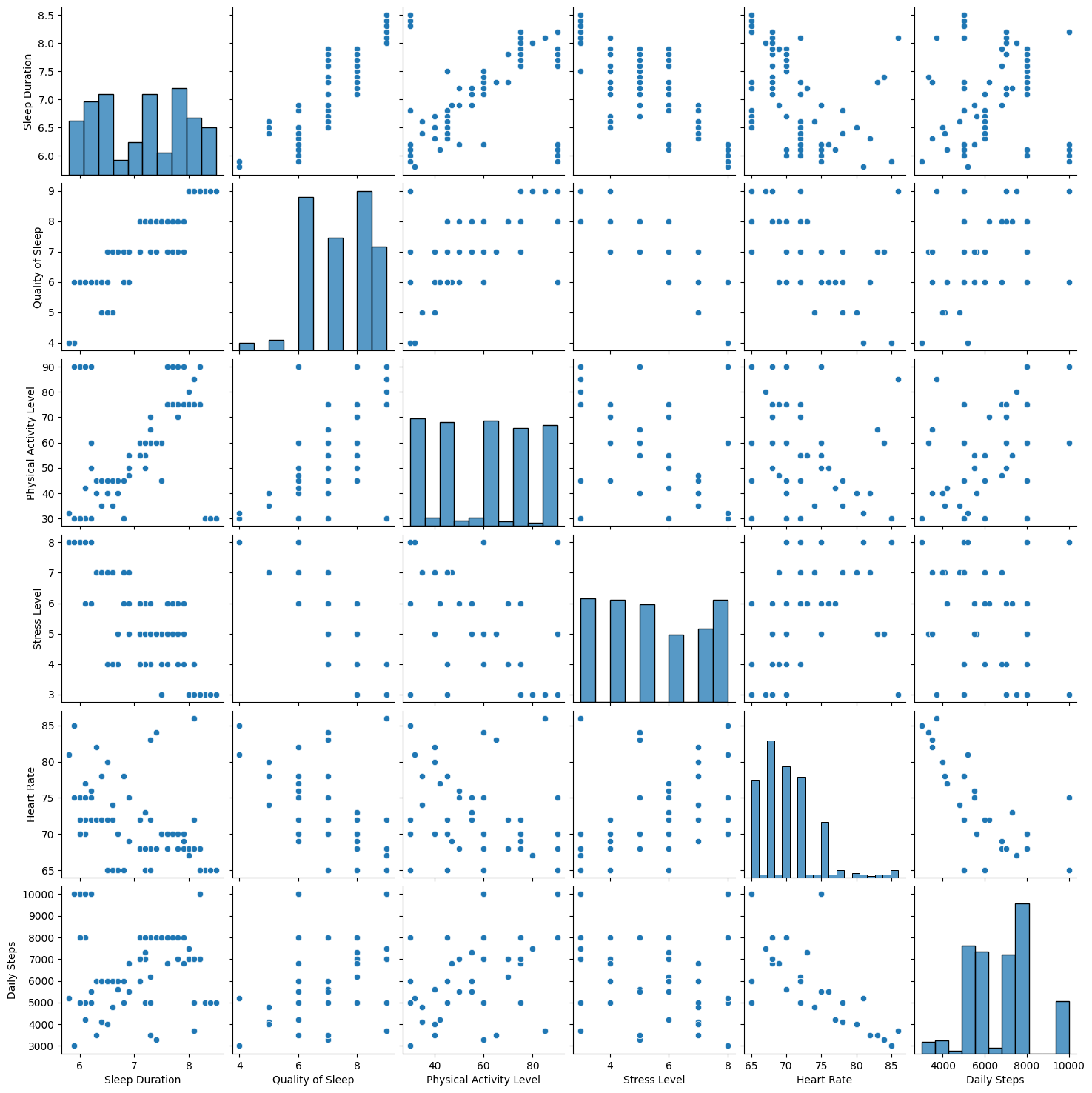
This visualization of pairplot provides a comprehensive overview of the relationships and distributions among these features.
df2= df[['Sleep Duration','Quality of Sleep', 'Physical Activity Level','Stress Level','Heart Rate','Daily Steps']]
sns.pairplot(df2)
<seaborn.axisgrid.PairGrid at 0x7f197cbf6a60>

Modeling#
Single-variable(1D) LinearRegression#
Let’s see if Sleep Duration correlates with Stress Level.
Using Altair, make a scatter plot with “Sleep Duration” on the x-axis and with “Stress Level” on the y-axis.
c1 = alt.Chart(df).mark_circle().encode(
x=alt.X("Stress Level", scale=alt.Scale(zero=False)),
y=alt.Y("Sleep Duration", scale=alt.Scale(zero=False))
)
c1
An downward trend can be seen in the graph. Let’s try fitting a linear regression.
X_train, X_test, y_train, y_test = train_test_split(df[["Stress Level"]], df["Sleep Duration"], test_size=0.2, random_state=42)
reg1 = LinearRegression()
reg1.fit(X_train,y_train)
LinearRegression()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LinearRegression()
Here are the coefficients and intercepts:
reg1.intercept_
9.053225447629982
reg1.coef_[0]
-0.3571879148875095
There appears to be a negative correlation between “Stress Level” and “Sleep Duration”.
Here is a line graph of the fitted line.
df["y_pred_linear"] = reg1.predict(df[["Stress Level"]])
c2 = alt.Chart(df).mark_line(color="red").encode(
x=alt.X("Stress Level", scale=alt.Scale(zero=False)),
y=alt.Y("y_pred_linear", scale=alt.Scale(zero=False))
).properties(
title="Sleep Duration VS Stress Level (1D)"
)
c1+c2
The resulting coefficients and the generated visualization show that “Stress Level” and “Sleep Duration” are negatively correlated. That is, as the stress level increases, the duration of sleep decreases.
Check accuracy
reg1.score(X_train,y_train)
0.6416287259643453
reg1.score(X_test,y_test)
0.7106694132038407
The score for training data and testing data are similar. Thus, the problem of overfitting does not exist.
Polynominal Regression#
Here, we try to use an extension of linear regression: the Polynominal Regression. Because it can usually capture complex relationships more accurately and improves prediction accuracy
X_train, X_test, y_train, y_test = train_test_split(df[["Stress Level"]], df["Sleep Duration"], test_size=0.2, random_state=42)
pipe = Pipeline(
[
('poly', PolynomialFeatures(degree=3, include_bias=False)),
('reg2', LinearRegression())
]
)
pipe.fit(X_train, y_train)
Pipeline(steps=[('poly', PolynomialFeatures(degree=3, include_bias=False)),
('reg2', LinearRegression())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('poly', PolynomialFeatures(degree=3, include_bias=False)),
('reg2', LinearRegression())])PolynomialFeatures(degree=3, include_bias=False)
LinearRegression()
Here are the coefficients and intercepts:
pipe['reg2'].coef_
array([-6.10996642, 1.14379135, -0.07117077])
pipe['reg2'].intercept_
18.060560122823077
Here is a line graph of the fitted line.
df["y_pred_poly"] = pipe.predict(df[["Stress Level"]])
c3 = alt.Chart(df).mark_line(color = "red").encode(
x = "Stress Level",
y = "y_pred_poly"
).properties(
title="Sleep Duration VS Stress Level (Poly)"
)
c1+c3
By generating a visualization of the graph, we can observe that the fitting line is no longer a straight line, it becomes a zigzag line as the data changes.
Check accuracy
pipe.score(X_train,y_train)
0.7321725237071466
pipe.score(X_test,y_test)
0.7642135932204126
The score for training data and testing data are similar. Thus, the problem of overfitting does not exist. Also, the accuracy of the prediction is improved over the previous results using the Single-variable(1D) LinearRegression method.
Multi-variable LinearRegression#
Now, we perform Multi-variable LinearRegression. Instead of a single input variable, we use three: "Quality of Sleep", "Physical Activity Level", "Stress Level". This model typically has higher predictive accuracy when multiple influential factors are included.
cols = ["Quality of Sleep","Physical Activity Level","Stress Level"]
X_train, X_test, y_train, y_test = train_test_split(df[cols], df["Sleep Duration"], test_size=0.2, random_state=42)
reg3 = LinearRegression()
reg3.fit(X_train,y_train)
LinearRegression()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LinearRegression()
Here are the coefficients and intercepts:
reg3.coef_
array([ 0.52995689, 0.00194484, -0.04022593])
reg3.coef_[0]
0.5299568895665259
Here is a line graph of the fitted line.
df["y_pred_logistic"] = reg3.predict(df[cols])
c4 = alt.Chart(df).mark_line(color="red").encode(
x=alt.X("Stress Level", scale=alt.Scale(zero=False)),
y=alt.Y("y_pred_linear", scale=alt.Scale(zero=False))
).properties(
title="Sleep Duration VS Stress Level (Logistics)"
)
c1+c4
Check accuracy
reg3.score(X_train,y_train)
0.7844910687954103
reg3.score(X_test,y_test)
0.7809667489067984
The score for training data and testing data are similar. Thus, the problem of overfitting does not exist. And, compared to Single-variable (1D) LinearRegression and Polynominal Regression, the prediction accuracy of this model is further improved.
Summary#
We built three models to predict "Sleep Duration": Single-variable(1D) LinearRegression, Polynominal Regression, Multi-variable The accuracy of the three models is 0.71, 0.76 and 0.78 respectively. Moreover, we can conclude that “Stress Level” and “Sleep Duration” are negatively correlated.
Q2: Prediction of Sleep Disorder#
Details about Sleep Disorder Column:
None: The individual does not exhibit any specific sleep disorder.Insomnia: The individual experiences difficulty falling asleep or staying asleep, leading to inadequate or poor-quality sleep.Sleep Apnea: The individual suffers from pauses in breathing during sleep, resulting in disrupted sleep patterns and potential health risks.
Decision Tree#
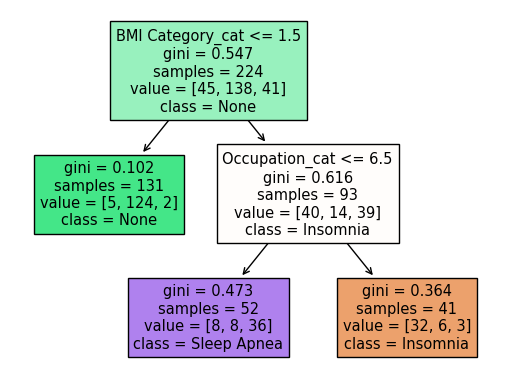
Train a model using the DecisionTreeClassifier to predict the category of sleep disorder based on an individual’s body mass index (BMI) category and occupation.
cols = ['BMI Category_cat','Occupation_cat']
X_train, X_test, y_train, y_test = train_test_split(df[cols], df['Sleep Disorder'], test_size=0.4, random_state=42)
clf1 = DecisionTreeClassifier(max_leaf_nodes=3, random_state=2)
clf1.fit(X_train,y_train)
DecisionTreeClassifier(max_leaf_nodes=3, random_state=2)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeClassifier(max_leaf_nodes=3, random_state=2)
Visualize decision trees to help understand how the model makes decisions.
import matplotlib.pyplot as plt
from sklearn.tree import plot_tree
fig = plt.figure()
_ = plot_tree(clf1,
feature_names=clf1.feature_names_in_,
class_names=clf1.classes_,
filled=True)
Check accuracy
clf1.score(X_train,y_train)
0.8571428571428571
clf1.score(X_test,y_test)
0.8666666666666667
The score for training data and testing data are similar. Thus, the problem of overfitting does not exist.
The decision boundary for our decision tree#
Here we use random number generator to generate (5000) random points for our chart
rng = np.random.default_rng()
arr = rng.random(size = (5000,2))
df3 = pd.DataFrame(arr, columns=cols)
df3['BMI Category_cat'] *= 3
df3['Occupation_cat'] *= 10
df3['pred'] = clf1.predict(df3[cols])
Now we make the chart. This shows a decision tree with three leaf nodes (three regions).
alt.Chart(df3).mark_circle().encode(
x = 'BMI Category_cat',
y = 'Occupation_cat',
color = 'pred'
)
Ramdom Forest#
In order to improve the accuracy of the prediction, we attempt to use Random Forest. By constructing multiple decision trees and averaging their predictions.
First, we add two corresponding variables: ‘Age’, ‘Stress Level’.
cols = ['Age','Stress Level','BMI Category_cat','Occupation_cat']
X_train, X_test, y_train, y_test = train_test_split(df[cols], df['Sleep Disorder'], test_size=0.4, random_state=42)
rfc1 = RandomForestClassifier(n_estimators=200, max_leaf_nodes=5)
rfc1.fit(X_train, y_train)
RandomForestClassifier(max_leaf_nodes=5, n_estimators=200)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
RandomForestClassifier(max_leaf_nodes=5, n_estimators=200)
Check which features are more important for prediction
pd.Series(rfc1.feature_importances_, index=rfc1.feature_names_in_).sort_values(ascending=False)
BMI Category_cat 0.366654
Occupation_cat 0.317599
Age 0.245178
Stress Level 0.070569
dtype: float64
Check accuracy
rfc1.score(X_train,y_train)
0.9107142857142857
rfc1.score(X_test,y_test)
0.8733333333333333
The results obtained are similar to the Decision Tree model, slightly better.
Summary#
Since “Sleep Disorder” is Categorical, we built two models, Decision Tree and Ramdom Forest, to predict it. Compared with Decision Tree, we added two corresponding variables in Ramdom Forest. But the accuracy difference between these two models is quite small. Their accuracy is 0.86 and 0.87 respectively.
Summary#
This project analyzed the influence of daily habits and lifestyles on sleep status, particularly sleep duration and sleep disorders, through multiple models. The accuracy of the models demonstrated the strong association between these factors and sleep quality. We found a negative correlation between stress levels and sleep duration. Furthermore, although the accuracy of the decision tree and random forest models was similar in predicting sleep disorders, this demonstrates the potential value of different modeling approaches in solving real-world problems.
References#
This dataset Sleep Health and Lifestyle Dataset was obtained from Kaggle.